Overview
As a powerful tool for the identification of complex trait-associated genes, transcriptome-wide association study (TWAS) has gained popularity over the years due to its ability to overcome the challenge of complex architecture of genetic regulation. However, because TWAS usually focuses on identifying associations between genes and diseases at the tissue level, translating these associations into actionable targets remains a significant challenge due to the heterogeneous cellular composition in the tissue microenvironment.
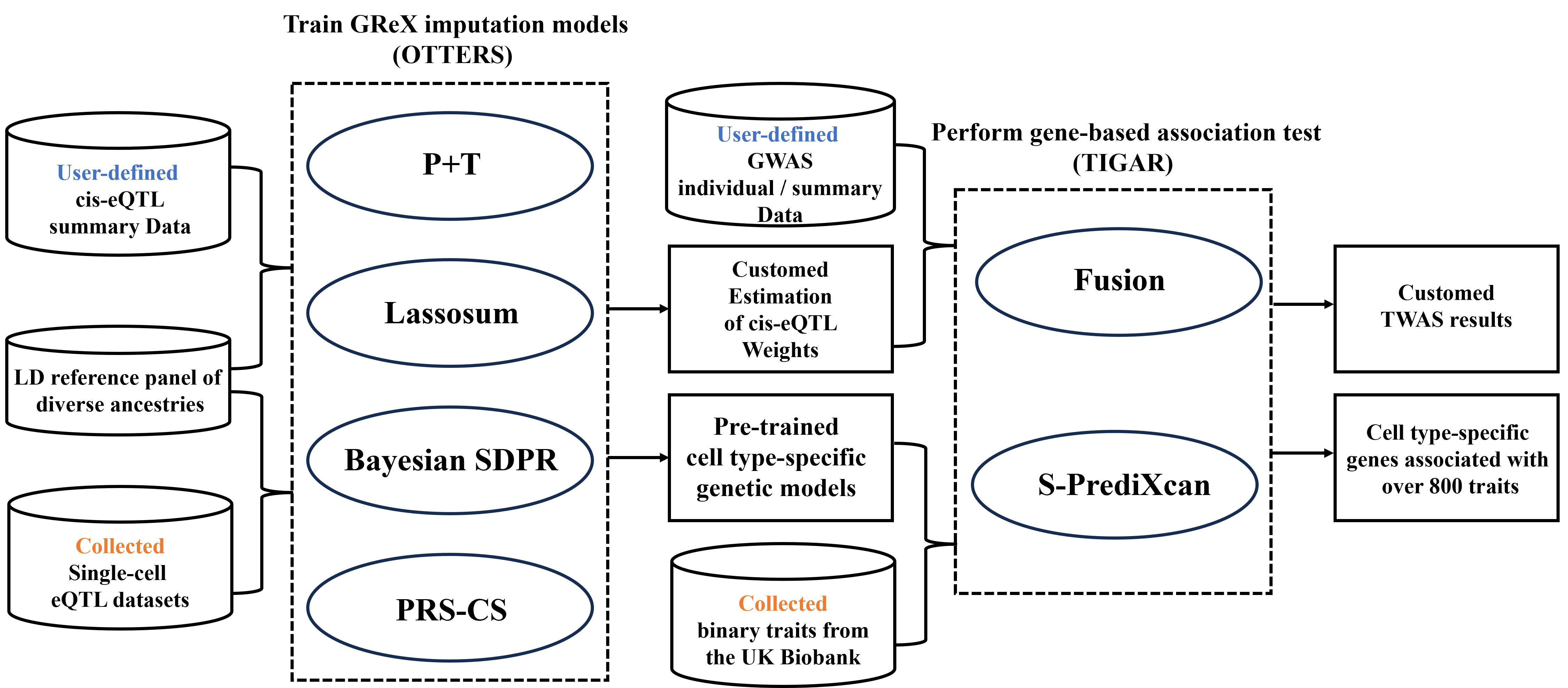
Here, we introduce a database for discovering trait-associated genes through single-cell transcriptome-wide association studies (scTWASdb), which provides a platform for training cell type-specific models for genetically regulated gene expression (GReX) and conducting gene-based association tests. Meawhile, scTWASdb also provides comprehensive resource for molecular and genetic associations with 829 binary traits sourced from the UK Biobank at single-cell resolution.
Query
Discover cell type-specific genes/SNPs associated with traits from the UK Biobank.
Association ListBrowser
Retrieve information for cell type-specific models, collected traits, and studies
Detailed informationTrain imputation models
Train customed genetic GReX models with uploaded eQTL summary data.
RunTraining